
A comprehensive solution to the challenge of siRNA Design
iRNAchek™ provides a comprehensive and intuitively designed environment for organization of sequence data, design of siRNAs, and tracking and analysis of successful templates.
OVERVIEW
Five great reasons to try OptGene™:
Intuitive Environment:
iRNAchek™ provides a comprehensive and intuitively designed environment for organization of sequence data, design of siRNAs, and tracking and analyzing successful templates.
Flexible Design:
Allows users flexibility to define their search templates, set regions of interest based on wide variety of criterion such as ORFs, UTR regions, introns and exons, secondary structures, motifs and domains, and post design analysis of successful siRNA.
Easy to learn:
User-friendly organization of features, GUIs and visualization tools which provide knowledge rather information.
Robust Analysis Capabilities:
iRNAchek™ has a unique tool that keeps track of successful templates and uses statistical techniques to convert information contained in siRNA fragments into knowledge.
24 X 5 Free Support:
Free Live web demonstration and post sale e-mail support for the life.
FEATURES
Search based on specific region
iRNAchek™ provides a facility to design the siRNA based on the regions of interest. These regions can be set based on Open Reading Frames, introns, exons, UTR regions and other sequence annotations. There is also a provision to eliminate templates from specific regions.
Motif Search
It is very essential to design siRNA targets that do not fall within functionally significant motifs and domains of the sequence. iRNAchek™ takes care of this efficiently by the use of a filter to refine the results based on Motif and Domain search.
Hairpin Loop search
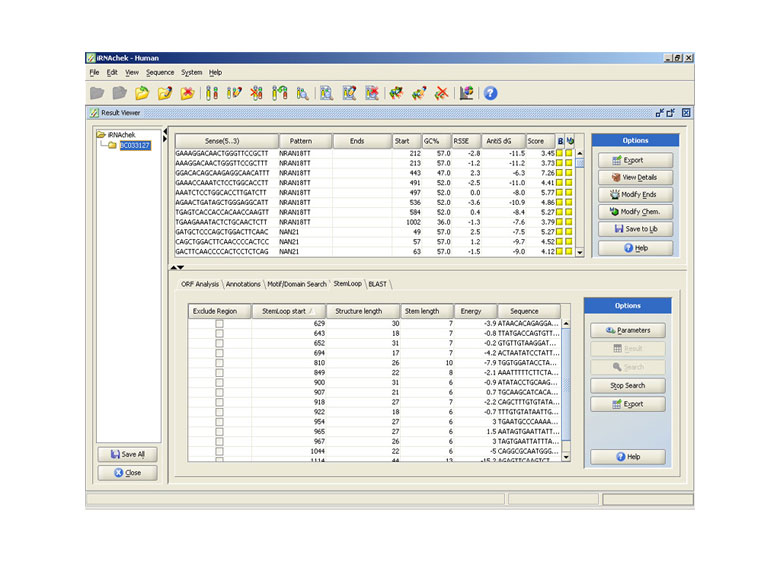
Efficiency of siRNA is sometimes affected by existence of stable hairpin loops. This tool identifies all possible hairpin loop formations and sorts them on the basis of their stabilization energy. User has an option to filter siRNA’s overlapping with selected hairpin loops.
Statistical Analysis
iRNAchek™ has a unique tool that keeps track of successful templates and uses statistical techniques to convert the information contained in siRNA fragments into knowledge. This tool helps the user to design targets with better
BLAST Tool
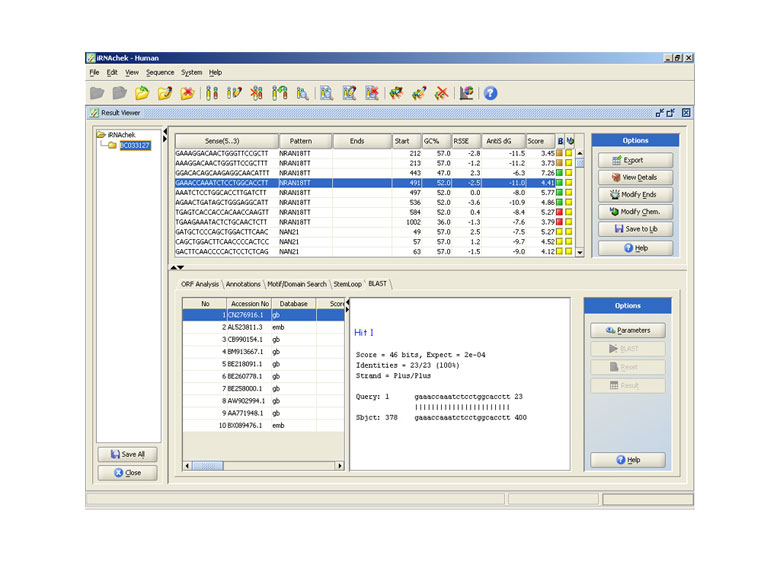
iRNAchek™ helps the user select specific siRNA’s using intuitively designed BLAST tool. Using this tool the user can search for potential matches from NCBI database or can create his own proprietary database and search for matches against proprietary databases for possible matches.
Chemical Modifications
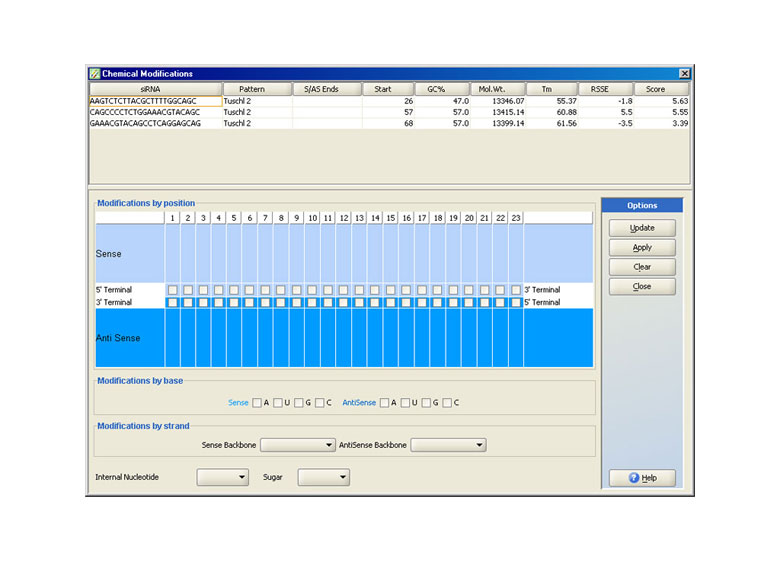
iRNAchek™ allows users to enhance siRNAs by allowing the user to define an implement chemical modification rules. The user can change overhang length, vary bases of overhangs to include deoxy bases, modify the molecule backbone, sugar groups, individual nucleotide bases including terminal bases and further. These modifications may be applied by position, by base, and by strand.
siRNA library
iRNAchek™ provides a user a library to store designed siRNA templates, along with any modification done. A set of siRNA selection filters, are given to the user for enabling performance analysis.
