
A comprehensive, multi-platform, sequence analysis software package
Genchek™ is a comprehensive, multi-platform, sequence analysis software package. It facilitates analysis of Expressed Sequence Tags (ESTs), Complete Genome, and SNP (Single Nucleotide Polymorphism) data.
Genchek™ is a research information and experiment management tool that integrates public and proprietary data through a discovery workspace that provides contextual access to sequence analysis tools, content and services. Genchek™ has an integral database system that can be used to access, store, organize and retrieve DNA, protein and vector sequences in an intuitive environment that offers editing, management and annotation of sequences.
OVERVIEW
Ten great reasons to try Genchek™:
Seamless Information Management:
A research information and experiment management tool integrating public and proprietary data through a discovery workspace that provides contextual access to sequence analysis tools, content and services.
Manage voluminous sequence data:
Integral database system to access, store, organize and retrieve DNA, protein and vector sequences in an intuitive environment that offers editing, management and annotation of sequences.
Robust Analysis Capabilities:
Manipulate and normalize raw data for better analysis, export superior graphics readily, thereby ensuring easy and timely access to pertinent information.
Multiple BLAST Capabilities:
Creating and managing databases and their sequences on any server made easier with Local BLAST Manager – can be used to apply powerful BLAST searches, even with a simple laptop.
Web Integration:
Connect to several public databases and utilities from the application to get holistic analysis results.
Platform Independence:
Can be run on any OS (Solaris, Mac, Windows , Unix/Linux)
Affordable Price:
ust pay once! Perpetual Licensing.
24 X 5 Free Support:
Free Live web demonstration and post sale e-mail support for the life.
Consultative approach of Development:
We continuously improve the software based on the latest scientific developments and your feedback.
Upgrade Price:
First year upgrade free. Minimal Upgrade fee thereafter and Dedicated support contract(Optional).
FEATURES
Integrated Sequence and Graphical Viewer
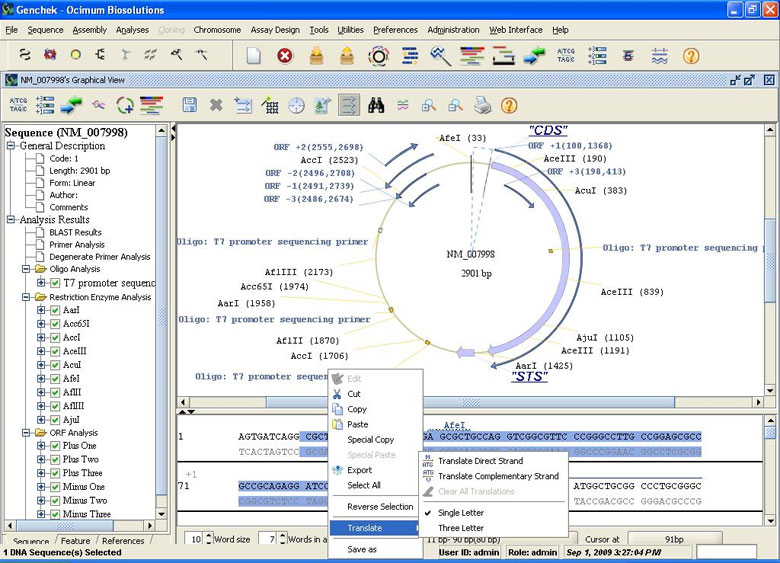
- Sequence as single strand, double strand, translation and with annotations
- Publication quality circular and linear sequence maps in graphical viewer
- Graphical view of restriction maps that facilitates easy cloning
- Graphical view of ORFs in the sequence maps
- Imported and user defined sequence annotations in graphical and sequence views
BLAST Suite
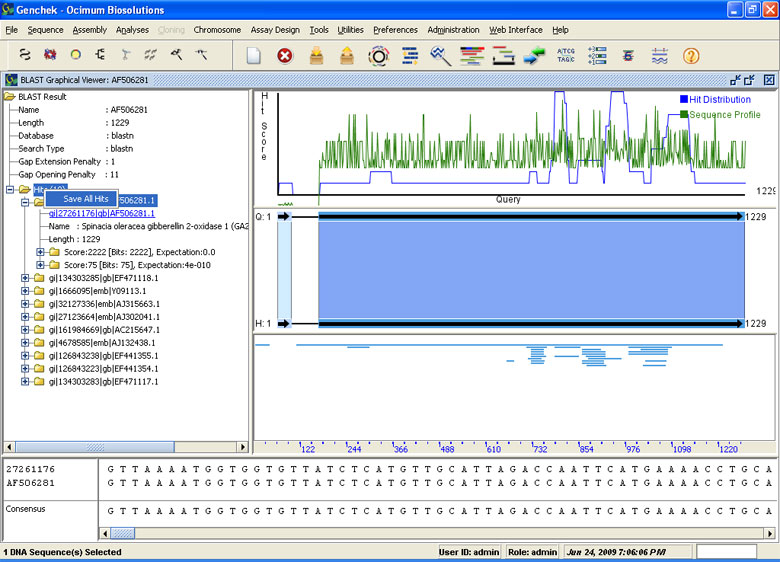
- Supports blastn, blastp, blastx, tblastn, tblastx and Batch Blast
- Create customized local BLAST databases through our pluggable local BLAST manager
- BLAST queries against multiple local databases
- Customized interface to search and view BLAST hits graphically
- Hyperlinked and downloadable accessions to import BLAST hits on a single click
- Importing of Multiple BLAST Hit Sequences
- Search dbSNP based on user defined parameters
Sequence Alignments
- Sequence Alignment Editor
- Align sequence pairs through Global and Local alignments
- Dot plots for possible alignments, and secondary structures
- Display alignment results using user-configurable color scheme
- Align multiple sequences
- Edit and save the alignments to a local database
- Save alignments in FASTA, PFAM, MSF, Clustal, BLC, PIR formats and associated trees in Newick format
- Analyze Phylogenetic relationships using neighbor joining algorithm and UPGMA
Electropherogram Viewer
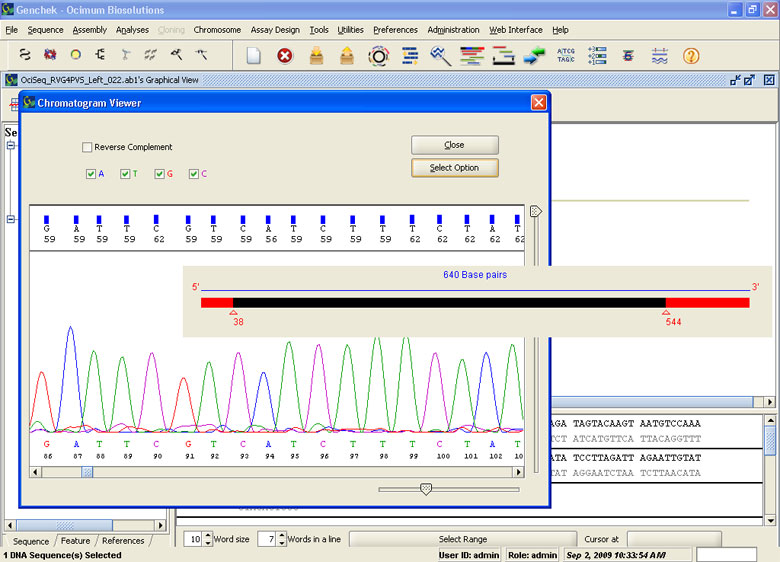
- Import all major chromatogram formats (e.g. .abi, .scf, .abd)
- Import, export and modify chromatograms
- Analyze the quality of sequence information
- Manipulate the chromatogram data for filtering weak signals
- Quality Values display in the Chromatogram Viewer
- Automatic End Clipping based on Quality Values
Contig Assembly
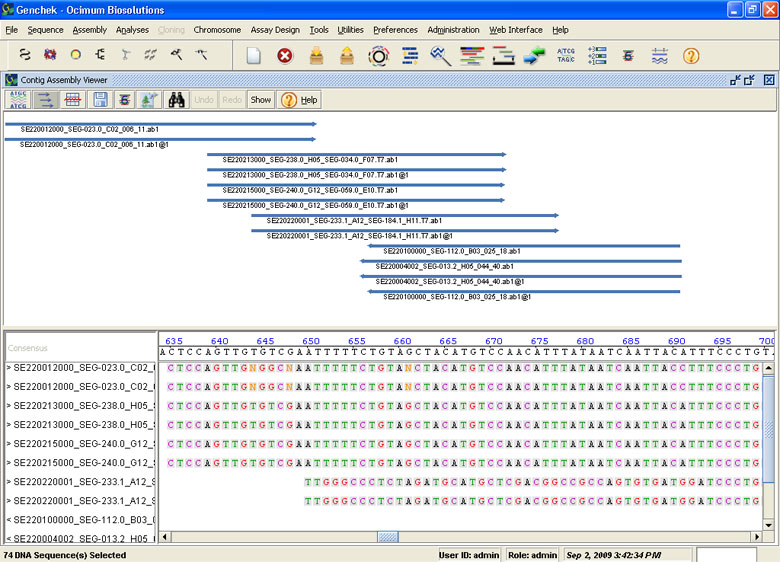
- Automatically align and assemble large numbers of sequences into contigs
- Allows overlap region length selection, fragment inversion check and identification of consensus regions
- ave contig results and export contig map
Automatic Vector Trimming
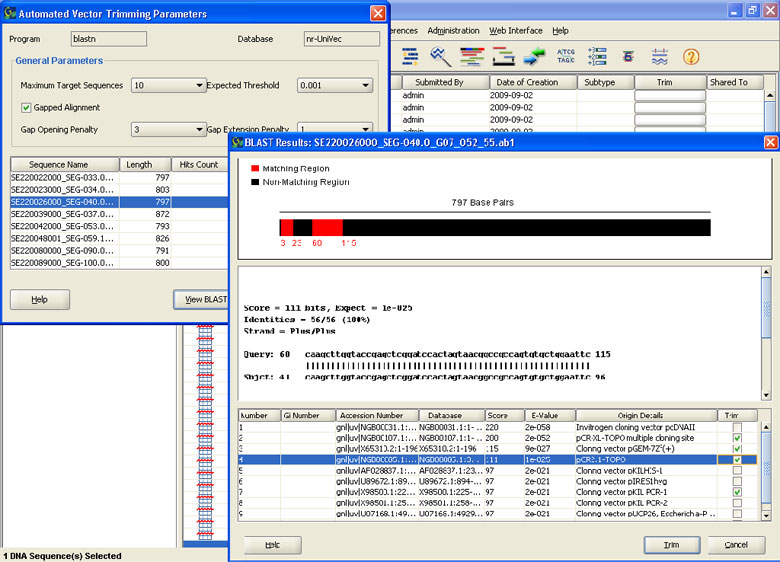
- Performs trimming in one step on multiple sequences
- Allows users to search against any sequence in a specialized non- redundant vector database using BLASTn, trim that region automatically
- Trimmed sequences information shown in graphical viewer with facility to print and export results
DNA Sequence Analysis
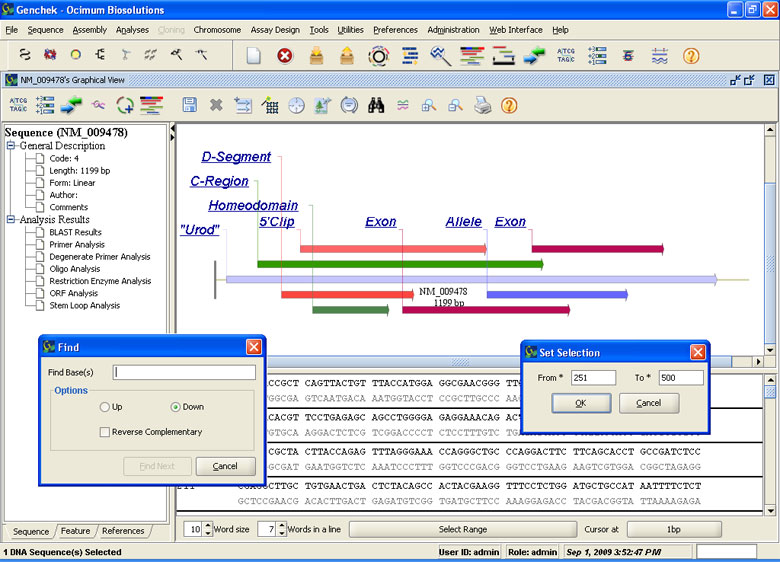
- Retrieve sequences in all standard sequence formats EMBL, Genbank, FASTA, DDBJ and popular sequence analysis software
- Feature information and annotations are loaded automatically
- Monitor the quality of imported sequence information
- Import chromatograms directly from sequencing machines
- Create and manipulate sequences using Sequence Editor
Feature Extraction Tool
- Allows for Feature Extraction on DNA/Vector/Protein sequences with facility to print and export results
Protein Sequence Analysis
- Reverse translation
- Protease digest
- Generate plots for Amino acid frequency, amino acid weights, Charge vs pH, and by Hydrophobicity
- View statistics for various physico-chemical properties of the protein sequence
- Analyze protein sequence for hydrophobicity, with helical wheel or beta staircase representations
Pattern Search
- Search for DNA patterns, motifs and domains
- Search across a sequence or entire experiments
- Create your own patterns and save them for future
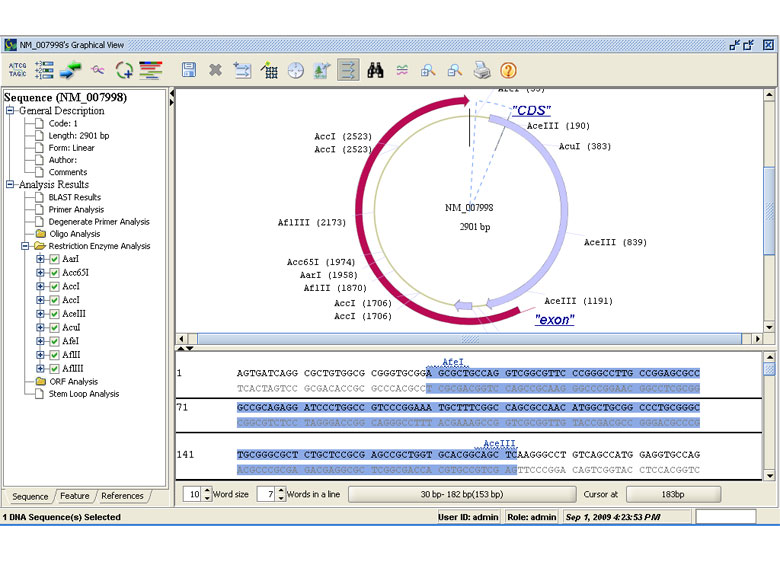
Restriction Analysis
- Simulate restriction analysis experiment
- Define custom enzyme sets
- View cut site and restriction information Graphical view of RE analysis for ease of work and comprehension
Primer Design
- Degenerate primer design to amplify homologous sequences
- Design optimal Primers for PCR and sequencing
- Design primers unique to entire sequence or to a selected region
- Analyze Primers for Secondary Structures
- Facility to export and save Primers for further analysis
TaqMan® Probe Design
- Performs TaqMan design with Secondary structure search and Cross Homology
- Probe will be searched for whole sequence length, 5′ biased or 3′ biased, search ranges of interest
- TaqMan Design allows import of Online Sequences and perform TaqMan Design on them
- Allows users to view TaqMan probes for single sequence or view all the found TaqMan for that sequences on result table
Exon Junction Primer Design
- Locates exon positions and number of exons in the sequences
- Primer pairs will be designed in the given amplicon range, either sense primer or antisense primer is designed on the exon junction with a facility to export results
SSR Marker Primer Design
- Facilitates searching SSR markers based on motif length range and their frequency
- Allows to perform SSR primer design on single DNA or vector sequence with a facility to export SSR and primer design results
Cloning Module
- Design new vectors with RE-based cloning supported by graphical interface of sequences
- Facilitates LR and BP reactions of Gateway cloning protocols
- Generates B-cassettes through PCR amplification
Data Management Capabilities
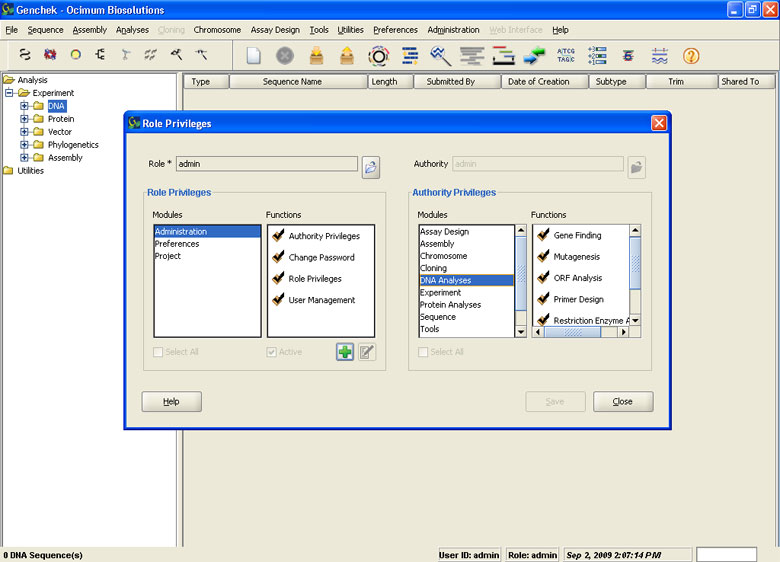
- Relational database support to organize experiment data
- Store and manage DNA, protein, vector sequences, Restriction Enzymes, Oligonucleotides and BLAST Results
- Client server architecture to share your data over an internal network
- Import and export data in all popular formats and share results with your colleagues
- Define user specific privileges to access data
Extensive Web Interface
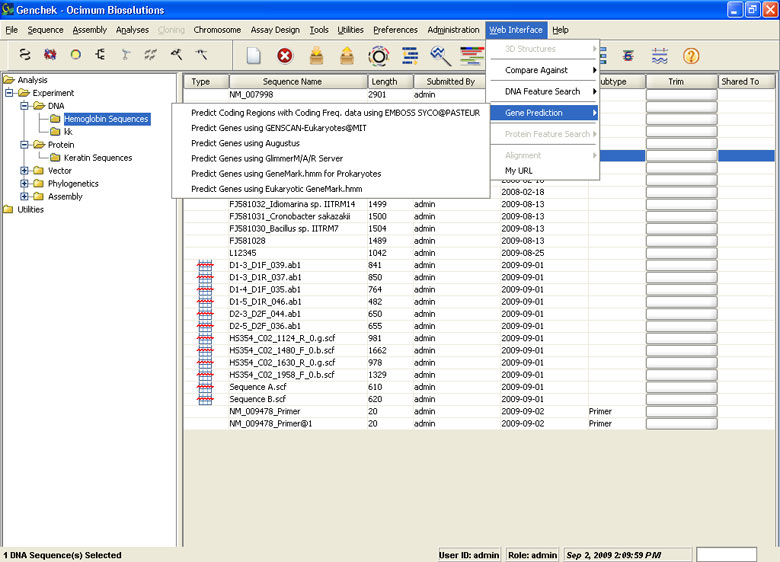
- Connect to several public database and utilities from the application
- Add, delete or edit your own set of regularly used URLs
Local BLAST Manager
- Create and manage databases and their sequences on any Server
- Apply the powerful BLAST searches against multiple databases using an ordinary desktop or laptop computer
Additional Features
- Renaming Multiple Sequences Simultaneously
- Importing Species Names along with the Sequence
- QTL Mapping
Technical Support
- Ocimum’s technical support staff is available 24 hours, five days a week, to answer your questions about Genchek™ over phone, e-mail and web chat.
